Globally; CMV has emerged as the most important
cause of congenital infection, in recent years. Congenital CMV infection
may lead to hearing, cognitive and motor impairment in babies [1,2].
The large CMV genome encodes several hyper variable
loci. Many genetically different strains of CMV circulate in the human
population. It has been suggested that difference in virulence,
pathogenicity, progression and severity of disease in immunocompromised
individuals, including transplant recipient may be attributed to
variation between HCMV strains. Conside-rable attention has recently
been focused on the analysis of strain variation among HCMV isolates.
Some 20 different strains have been isolated and differentiated by
restriction analysis of PCR amplified DNA fragments [3-5]
Glycoprotein B gene of Cytomegalovirus plays an
important role in virus infectivity, cell to cell spread and is a major
target for antibody mediated immunity [6,7]. There are several variable
regions of gB and CMV strains have been classified into four genotypes
based on restriction fragment length polymorphism(RFLP) of a fragment
corresponding to the cleavage site between residue 460 and 461 [5,8].
These genotypes have been reported to be clinically associated with the
outcome of the disease though these reports are contradictory [9-12].
Recently evidence of new strains arising in individuals infected with
multiple CMV strains and intragenic variations within gB gene were
obtained [9,10].
Few sero-epidemiological studies conducted in Indian
population, have shown that there is prevalence of 80-97% seropositivity
of CMV antibodies (IgG) in women of child bearing age [13-15]. High
incidence of congenital CMV infection in babies due to in-utero CMV
infection in mothers by reactivation/reinfection or primary infection of
the virus has also been reported besides perinatal infections acquired
by newborns [15-18]. The magnitude of the problem, in India has not been
systematically investigated. The present study was undertaken to
determine the prevalence of congenital CMV infection in symptomatic
babies and the gB genotyping of CMV strains circulating in the affected
babies. An attempt was made to associate the possibility of clinical and
prognostic significance of the prevailing genotypes.
Methods
Three hundred seventy five clinical samples (blood
and urine) from 375 infants (newborn to 6 months old) babies exhibiting
various symptoms of congenital infection, referred to Virology
laboratory of the National Centre for Disease Control were selected for
the present study. The samples were accompanied by duly-filled proforma
with relevant history and clinical details . All the samples were
negative for HIV, and without any history of blood/blood product
transfusion. Blood and urine samples from 60 asymptomatic babies,
referred to NCDC for other testing were included in the study and
treated as control group.
Blood samples were clotted and centrifuged for serum
separation prior to testing. All the sera were stored at -20ºC pending
testing. Serum samples of the babies were tested for CMV-IgM antibodies
using µ-capture ELISA (RADIM). DNA was isolated from urine samples and
serum samples by QIamp DNA Mini kit (QIAGEN, Germany) as per
manufacturer’s protocol. Fragment from gB (UL55) gene region was
amplified from isolated DNA samples using pre-published DNA
oligonucleotide primers [19], by nested PCR. Primers for Outer-PCR: gB-1
CAAGARGTGAACATGTCCGA, gB-2 GTCACGCAGC TGGCCAG. Thermal Cycling
Profile: Initial Denaturation 95ºC: 10 min; Amplification:35 Cycles:
Denaturation 95ºC : 1 min; Annealing 55°C 1min ; Extension 72ºC:1 min;
Final Extension 72ºC:7 min.; Inner-PCR: gB-3 TGGAAC
TGGAACGTTTGGC, gB-4 GAAACGCGCGGC AATCGG. Thermal Cycling Profile:
Initial Denaturation 95ºC 10 min; Amplification: 35 Cycles: Denaturation
95
ºC: 30 seconds; Annealing:
54ºC: 45seconds; Extension 72ºC: 30seconds; Final Extension 72°C: 7 min.
The PCR products (520bp, 305bp) were subjected to gel
electrophoresis on 1.5% agarose to visualize the product with ethydium
bromide stain. The inner amplified products from gB gene region were
digested with the Restriction Enzymes, Hinf1 and Rsal (Promega Madison,
Wise, USA). Approximately 1 µg of the PCR product was added to 2 µL of
PCR buffer and 1unit of enzyme, mixed gently, spinned briefly and
incubated at 37
ºC for 3 hrs.
for digestion. The digested fragments were analyzed on 3% agarose gel.
The 50 bp DNA ladder was also loaded on gel to compare the fragment
length of the digested products. Distinct gB genotypes could be
identified by the different lengths of restriction fragments (Table
I). Sequencing was also carried out from the inner (305bp) PCR
product using Big Dye Terminator cycle sequencing ready reaction kit
(Applied Biosystems, USA).
TABLE I Fragment Length (BP) Of Four Gb Genotypes Identified By Rflp Analysis With Hinf1 Or Rsal Digestion
|
gB-1 |
gB-2 |
gB-3 |
gB-4 |
|
HinfI |
202,67,36 |
202,100 |
202,97 |
202,67,36 |
|
Rsal |
239,66 |
239,63 |
195,63,41 |
195,66,44 |
Representative sequences derived from this study were
submitted to Genbank at NCBI website and accession numbers acquired.
Bioinformatics software viz. BLAST, CLUSTALW, BIOEDIT, MEGA v.4 were
used for sequence analysis.
Results
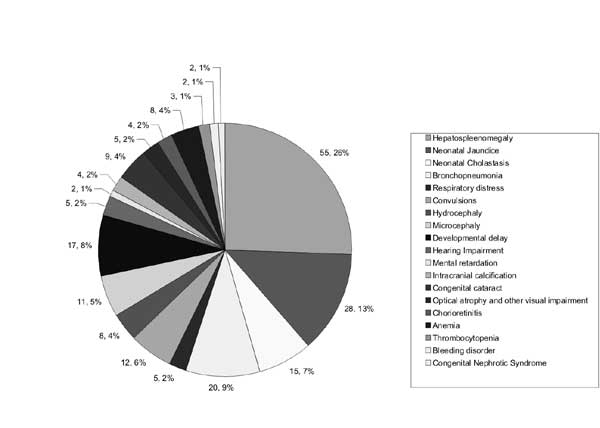
Among the clinical manifestations displayed by the
babies, Hepatosplenomegaly was the most common feature (55.3%) followed
by neonatal jaundice, broncho-pneumonia, developmental delay,
neonatal cholestasis, micocephaly and congenital cataract. Hearing
impairment, hydrocephaly, and chorioretinitis were also present in some
infants (Fig. 1).
 |
|
Fig. 1 Clinical features in babies
with congenital/perinatal CMV Infection.
|
The high incidence of congenital CMV infection
(19.4%) was detected among babies born with various birth defects. The
maximum number of symptomatic babies presented to the hospitals in the
age group of (O-1M) and the case reporting decreased in the subsequent
months. Positivity rate was highest in the age group (1+ to 2 months)
(28%) (Table II). Higher number of male infants (64%) were
positive. All the samples of babies from the control group were negative
for congenital CMV infection by ELISA and PCR.
TABLE II Infants Positive For Anti Cmv-igm Antibodies (N=75)
|
Age group(mo) |
No. of babies |
Total (%) |
|
Male |
Female |
|
|
NB-1 |
6 |
4 |
10 (13.3%) |
|
1 + to 2 |
15 |
6 |
21 (28%) |
|
2 + to 3 |
13 |
7 |
20 (26%) |
|
3 + to 4 |
9 |
6 |
15 (20%) |
|
4 + to 5 |
3 |
2 |
5 (6.67%) |
|
5 + to 6 |
2 |
2 |
4 (5.33%) |
|
Male : Female 48 (64%) : 27 (36%) |
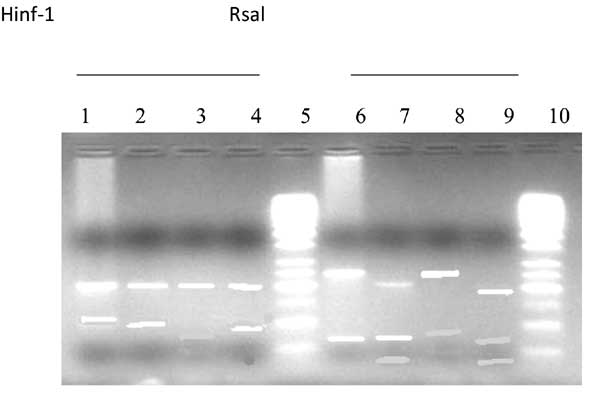
RFLP of gB (UL-55) PCR products for genotyping
demonstrated prevalence of gB [1-3] genotypes in the babies. gB 4 was
not detected (Fig. 2). The frequency of gB3 was the most
dominant (49.25%) followed by gB1 (24.4%) and gB2 (22.4%).
 |
|
Fig. 2 Restriction enzyme analysis of
gB genotype by Hinf1 & Rsal. Lane-1,6: gB-2; Lane-2,4,7,9 :
gB-3; Lane-3,8 : gB-1; Lane-5,10: Molecular markers of 50 bp
|
GenBank accession numbers of the representative
submitted sequences for gB gene region from this study are:
EU938342 to EU938348; HM069142 to HM069157. Sequences from variable
region of gB gene (cod 448 to 480) including proteolytic cleavage site
were compared to the published sequences of 4 gB genotypes. The result
from the sequencing was concordant with RFLP results.
The prevailing gB genotypes and specific CMV disease
manifestations in infants were compiled to analyse the existence of
correlation with pathogenesis of the disease. It was found out that
liver disorders were mainly associated with gB3 genotype, and hearing
impairment and central nervous system symptoms with genotype gB2 (Table
III).
TABLE III gB Genotypes Distribution in 67 Babies Who Were Positive For Congenital/Perinatal CMV Infection
|
gB genotypes/ Clinical manifestations
|
gB-1 |
gB-2 |
gB-3 |
Failure |
|
Hepatosplenomegaly, NNcholestasis, jaundice, pneumonia and
respiratory distress, developmental delay with other clinical
features like anemia, thrombocytopenia
|
9 |
1 |
26 |
-
|
|
Clinical features involving CNS and long term seuelae (Microcephaly,
hydrocephaly, hearing impairment, intracranial calcification,
mental retardation etc.) with other secondary clinical features
of respiratory disorders. |
1 |
10 |
2 |
- |
|
Congenital cataract, chorioretinitis, optical atrophy and other
visual impairments with many other anomalies like developmental
delay, hepatospleenomegaly,anemia,pneumonia etc. |
7 |
4 |
5 |
2 |
|
Total |
17 |
15 |
33 |
2 |
Discussion
A high incidence of congenital CMV infection (19.4%)
was detected among babies born with various birth defects, similar to
our previous study and other studies conducted in India and globally
[15-17,20,21]. The higher number of positive cases in the age group (1+
to 2 months) (28%), could be due to the fact that excretion of CMV virus
and antibody development is mostly detected after 2-3 weeks of birth in
congenital CMV infection besides cases of perinatal infection.
In the present study clinical samples from the
patients were processed for diagnosis and molecular studies to avoid any
variations which might occur during cell culture and subcultures.
Clinical samples were sufficient to carry out testing and retesting of
the samples.
RFLP of gB (UL-55) PCR products for genotyping and
sequencing showed that only 3 genotypes (gB1, gB2 and gB3) were
prevalent in the babies. Some studies from other countries have also
reported prevalence of these 3 genotypes in infants with congenital CMV
infections [22, 23]. No data on gB genotyping in babies with congenital
CMV infection is available from India though this kind of study in
immunocompromised patients have been conducted earlier [24, 25], in
which the prevalence of all 4 genotypes and mixed infection of gB
genotypes has been documented. No mixed infection was seen in this
study, but in two digested PCR products, the fragment pattern could not
be identified. Among the prevailing genotypes the frequency of gB3 was
the most dominant (49.25%). The findings from the present study
are different from some other studies from China and South Hungary,
where they have found gB1 as the most dominant genotype circulating in
babies with congenital CMV infection [23, 26, 27]. Various clinical
studies have suggested that gB genotype of HCMV strains may influence
the clinical outcome of acquired CMV infection [9-11]. Earlier global
studies have shown that gB1 genotype is associated with
hepatospleenomegaly [23,26]. The new feature demonstrated by the present
study was that babies who had manifestation of hearing impairment and
symptoms with CNS association had mainly genotype gB2 infection. It has
not been reported in any other study done on congenital CMV infection.
However, this finding needs to be explored further as the sample size
was small in this study.
To date, most knowledge regarding the medical
implications of viral disease stems from studies of acute viral
infections. Symptomatic congenital CMV infection is a significant cause
of morbidity in developing as well developed countries like United
States. It has been estimated that direct and indirect costs for
treating babies with congenital CMV infection approaches $1 billion
dollars per year[28]. As a consequence, prevention of this disease has
become a global issue for vaccine development, particularly for
administration to seronegative susceptible women of child-bearing age
[29, 30]. A virus such as CMV, which is able to establish latency and
evade immune surveillance, presents particular challenges in the
development of effective vaccination as HCMV genome displays great
genetic heterogeneity. Thus, there is a constant need for upgrading the
information on molecular epidemiology of CMV in different population,
which would help in forming efficacious vaccines for the prophylactic
treatment of CMV in humans.
Glycoprotein B (gB) is a major target for
neutralizing antibodies and an important component of recombinant
vaccines, under trial. Therefore, more studies on genetic variability
data in gB gene, from India may help to determine the optimal strains
for vaccine development in Indian population
Acknowledgments: Support from the Delhi
based government hospitals is duly acknowledged for providing the
clinical samples from symptomatic infants. We thank for the technical
support from staff of Microbiology, NCDC, Delhi.
Funding: None; Competing interests: None
stated.
References
1. Friedman S, Ford-Jones EL. Congenital
cytomegalovirus infection: an update. Pediatr Child Health.1999;4:35-8.
2. Benjamin Bar-Oz, Berkovitch M, Lee Ford-Jones,
Koren G. Congenital cytomegalovirus infection: Is there a breakthrough?
Canadian Family Physician. 2001;47: 1179-81.
3. Grillner L, Ahlfors K, Ivarsson SA, Harris S,
Svanberg L. Endonuclease cleavage pattern of cytomegalovirus DNA of
strains isolated from congenitally infeced infants with neurologic
sequelae. Pediatrics. 1988;81:27-30.
4. Peckham CS, Garrett AJ, Chin KS, Preece PM, Nelson
DB, Warren DE. Restriction enzyme analysis of cytomegalovirus DNA to
study transmission of infection. J Clin Pathol. 1986;39:318-24.
5. Chou S. Differentiation of cytomegalovirus strains
by restriction analysis of DNA sequences amplified from clinical
specimens. J Infect Dis. 1990;162:738-42.
6. Britt WJ, March M. Human cytomegalovirus proteins.
Intervirology. 1996:39:401-12
7. Navarro D, Lannette C, Tugizov S, Pereira L.
Humoral immune response to Functional Regions of human cytomegalovirus
Glycoprotein BJ Med Virol. 1997;52:451-9.
8. Chou SW, Denninson KM. Analysis of interstrain
variation in cytomegalovirus glycoprotein B sequences encoding
neutralization-related epitopes. J Infect Dis.1991; 163:1229-34.
9. Shepp DH, Match ME, Ashraf AB, Lipson SM, Millan
C, Pergolizzi R. Cytomegalovirus glycoprotein B groups associated with
retinitis in AIDS. J Inf Dis. 1996;174: 184-7.
10. Trincado DE, Scott GM, White PA, Hunt C,
Rasmussen l, Rawlinson WD. Human cytomegalovirus strains associated with
congenital and perinatal infections. J Med Virol 2000;61:481-7.
11. Fries BC, Chou S, Boeckh M, Torok-Storb B.
Frequency distribution of cytomegalovirus envelope glycoprotein
genotypes in bone marrow transplant recipients. J Infect Dis.
1994;169:769-74.
12. Vogelberg C, Meyer-Koni U, Hufert FT, Kirste G,
Von Laer D. Human Cytomegalovirus glycoprotein B genotype in renal
transplant recipients. J Med Viro. 1996;50:31-34.
13. Rai A, Kumari S, Khare S, Gandhoke I., Bhatia R,
Datta KK. Maternal viral infections and their implications in congenital
defects of new borns. J Basic and Applied Biomedicine.
1995;3:1-9.
14. Jindal S, Aggarwal N. A pilot seroepidemiological
study of cytomegalovirus infection in women of child bearing age. Indian
J Med Microbiol. 2005;23:34-6.
15. Chakravarty A, Kashyap B, Rathi K. The
seroepidemiological study on cytomegalovirus in women of child-bearing
age with special reference to pregnancy and maternal-fetal transmission.
Indian J Pathol Microbiol. 2005;48:518-21.
16. Abraham M, Abraham P, Jana AK, Kuruvilla KA,
Cherian T, Moses PD, Mathai E, .John TJ, Sridharan G. Serology in
congenital infections: experience in selected symptomatic infants.
Indian J Pediatr. 1999;36:697-700.
17. Gandhoke I, Aggarwal R, Lal S, Khare S.
Congenital CMV infection in symptomatic infants in Delhi and surrounding
areas. Indian Journal of Pediatrics. 2006;73:1095-7.
18. Rekha S, Chandrasekhra MK, Yeshwanth M.
Cytomegalovirus infection acquired through blood transfusions. Indian
Pediatr. 1995;32:575-7.
19. Aldo De Albuquerque Cunha, Lauro Juliano Marin,
Victor Hugo Aquino, Luiz Tadeu Moraes Figueiredo. Diagnosis of
cytomegalovirus infections by qualitative and quantitative pcr in hiv
infected patients. Rev Inst Med Trop S Paulo. 2002;44:127-32.
20. Ho M. Cytomagalovirus. In: Mandell GL,
Douglas RG, Bennett JE. (Eds.) Principles and Practice of
Infectious Diseases. Third Edition.Chuchill Livingstone, New York, 1990,
p. 1159-72.
21. Ahlfors K, Ivarsson SA, Harris S. Svanberg L,
Holmqvist R, Lenmark B, et al. Congenital cytomegalovirus
infection and disease in Sweden and the relative importance of primary
and secondary maternal infections. Scand J Infect Dis.
1984;16:129-38.
22. Ahumada-Ruiz S, Taylor-Castillo S, Visona K,
Luftig RB, Herrero-Uribe L. Determination of human cytomegalovirus
genetic diversity in different Patient Populations in Costa Rica. Rev
Inst Med Trop S Palo. 2004;46:87-92.
23. Zhong Sheng, Chao Chun Zou, Ji Yan Zheng, Yan
Zhao. Cytomegalovirus gB genotypes and clinical features in Chinese
Infants with Congenital Infections. Intervirology. 2006; 49:281-5.
24. Sowmya P, Dhanya V, Madhavan HN, Therese KL.
Comparative efficacy of PCR-based restriction fragment length
polymorphism (RFLP) and multiplex PCR for glycoprotein B (gB) genotyping
of human cytomegalovirus. Indian J Med Res. 2007;126:122-7.
25. Novak Z, Ross SA, Patro RK, Pati SK, Kumbla RA,
Brice S, et al. Cytomegalovirus strain diversity in seropositive
women. J Clinical Microb 2008; 882-6
26. Lukácsi A, Tarodi B, Endreffy E, Bábinszki A, Pál
A, Pusztai R, et al. Human cytomegalovirus gB genotype 1 is
dominant in congenital infections in South Hungary. J Med Virol.
2001;65:537-42.
27. Barbi M, Binda S, Carropo S, Primache V, Didò P,
Guidotti P, et al. CMV gB genotypes and outcome of vertical
transmission: Studies on dried blood spots of congenitally infected
babies. J Clin Virol. 2001;21:75-9.
28. Manning FJ, Swaetz M (Eds.) Clinical
Trials. Review of the fialuridine (FIAU) clinical trials: National
Academy Press. 1995.
29. Gonczol E and Plotkin S. Development of a
cytomegalovirus vaccine: lessons from recent clinical trials. Exp Opin
Biol Therap. 2001;1:401-12.
30. Pas RE, Burke RL Development of Cytomegalovirus
Vaccines: prospects for prevention of congenital CMV infection. Semn
Pediatr Infect Dis. 2002;13:196-204.